This program analyses the read depth in a bam file of short read sequence data mapped to a genome assembly, and tries to identify regions that have unusual read depth and so might correspond to copy number variations.
To use CNVnator, you must specify a 'bin size', or window size. CNVnator then divides the genome assembly into nonoverlapping bins (windows) of this size, and uses the count of mapped reads in each window as the read depth for that window. In the CNVnator paper, they use 100-bp bins (windows) to analyse data from a trio from the 1000 Genomes Project.
In the case of the 1000 Genomes Project, the reads are from an individual sequenced as part of the 1000 Genomes Project, and are mapped to the human reference genome. In my case, I am using CNVnator to look at reads from a particular species, mapped to the assembly for that species, where we suspect that there are uncollapsed haplotypes in the genome assembly, which should be detected by CNVnator as putative 'deletions' (regions of read depth < 1). It can also find potential 'duplications' (regions of read depth > 1), which could be due to collapsed repeats. The CNVnator paper says that it is harder to find potential duplications than potential deletions, and I find that in practice it tends to identify fewer potential duplications than deletions.
In the CNVnator paper, they say that CNVnator can identify CNVs from a few 100 bases to megabases in length. Furthermore, the precision is good: 200 bp for 90% of the breakpoints in a test case studied in the CNVnator paper (using a bin size of 100 bp). The higher the coverage you have, the smaller the bin size you can use, which will give you greater precision. They recommend to use ~100-bp bins for 20-30x coverage, ~500-bp bins for 4-6x coverage, and ~30-bp bins for 100x coverage. However, they say that the bin size used shouldn't be shorter than the read length in your data.
Running CNVnator
1) Download the program
You can download CNVnator from the CNVnator download page.
2) Set environmental variables
Before running CNVnator you need to set the ROOTSYS variable to the directory where you unpacked the root distribution, eg.:
> setenv ROOTSYS "/nfs/users/nfs_b/bf3/bin/root/"
You then need to set the LD_LIBRARY_PATH variable:
> setenv LD_LIBRARY_PATH ${LD_LIBRARY_PATH}:${ROOTSYS}/lib
3) Preparing input data & preprocessing the data
The input files required for CNVnator are:
(i) a fasta file of scaffolds/chromosomes,
(ii) a bam file,
(iii) individual fasta files for each scaffold/chromosome.
I find that CNVnator needs you to have individual fasta files for each scaffold/chromosome in the current directory. As well as this, CNVnator seems to stall if the fasta files have a lot of characters per line of sequence (eg. 1000s of characters), but this was solved by reformatting my fasta files so that they had just 60 characters per line of sequence (using my script reformat_fasta.pl).
To run CNVnator, you must run a pre-processing step to pull the reads out of the bam file:
> /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -tree out.sorted.markdup.bam
where /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/ is the path to where you have installed CNVnator.
The other arguments are:
-root: the name of a file that CNVnator will make to store a pre-processed form of your data,
-genome: your genome assembly file,
-chrom: the chromosome/scaffold that you are interested in,
-tree: your bam file.
This step seemed to require quite a lot of memory (RAM). For a 419 Mbyte RAM file (6,138,830 100-bp reads), this needed 1.5 Gbyte of RAM. It took about 25 minutes to run.
4) Making a histogram
The next step is to make a histogram (presumably a histogram of the read depth, for all the windows in your genome assembly). This is not memory consuming. To run this you need to have fasta files in your current directory with the sequences of all your scaffolds/chromosomes. For example, if you have scaffolds scaffold1, scaffold2, etc., you will need files scaffold1.fa, scaffold2.fa, etc. in your current directory. You can then run:
> /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -his 100
The '100' at the end is the bin size (window size) that you want to use.
This step takes less than 1 minute to run.
5) Calculating statistics
I think this step is to calculate statistical significances (p-values) for the windows that have unusual read depth.
% /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -stat 100
Here the '100' at the end is the bin size (window size).
This step only takes a few seconds.
6) Partitioning
This step partitions the chromosomes/scaffolds into long regions (each one of which could be longer than the window size) that have similar read depth, and so presumably similar copy number.
> /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -partition 100
Again, the '100' at the end is the bin size (window size).
The README file for the CNVnator program says that this is the most time-consuming step.
It took just a few seconds to run in my case (on one scaffold).
7) Identifying CNVs
This step finds potential CNVs.
> /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -call 100 > out.100.cnvnator
Here the '100' is the window size. I have redirected the output to the file 'out.100.cnvnator'.
Again, this just took a few seconds to run.
According to the CNVnator README file, the columns of the output file are:
CNV_type coordinates CNV_size normalized_RD p-val1 p-val2 p-val3 p-val4 q0
normalized_RD: normalized to 1.
p-val1: is calculated using t-test statistics.
p-val2: is from probability of RD values within the region to be in the tails of gaussian distribution describing frequencies of RD values in bins.
p-val3: same as p-val1 but for the middle of CNV
p-val4: same as p-val2 but for the middle of CNV
q0: fraction of reads mapped with q0 quality
Strangely, I find that some of the p-values in the output file are greater than 1, which they obviously shouldn't be if they are really p-values. I'm not sure why this is..
8) Visualising a CNV region
You can visualise one of the CNV regions by typing (you will need to have logged in using 'ssh -X' if you are doing this on the Sanger farm):
[Note to self: I found this worked for me on pcs4, but not on the farm head nodes]
> /nfs/users/nfs_b/bf3/bin/CNVnator_v0.2.7/src/cnvnator -root out.root -genome ref.fa -chrom SVE.scaffold.00029.709875_77520_542822 -view 100
This will bring up a prompt '>'. Type the region you are interested in on the prompt, eg.:
> SVE.scaffold.00029.709875_77520_542822:1-10000
This will bring up a lovely picture of your region:
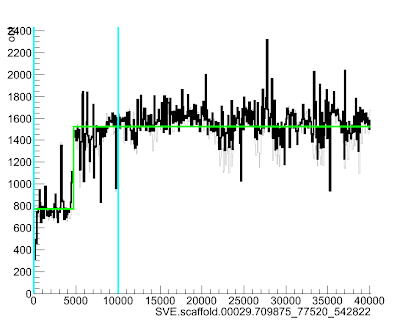
Here the green line goes up suddenly at position ~4700, suggesting that the read depth (and so copy number) increases quite a lot at that point in the scaffold.
Thanks to Bernardo Foth for a lot of help and instructions for running CNVnator (most of the above was worked out by Bernardo), and to Alan Tracey for example data.
A wrapper script for running CNVnator
It takes a bit of time to paste in all the above commands, so I've written a simple wrapper script to run CNVnator on an assembly, run_cnvnator_on_assembly.pl.
Running CNVnator on the Sanger compute farm
This will be useful to Sanger people only: if you want to run CNVnator on an assembly relatively quickly, it is useful to divide up your assembly into smaller subsets, and run each subset on a separate farm node. I have a script to do this, run_cnvnator_on_farm_wrapper.pl. It submits the jobs as a LSF 'job group', so that only 100 jobs run at any one time. For each job, it requests 3.5 Gbyte of RAM.
[Note to self: Latest cnvnator : /nfs/users/nfs_b/bf3/bin/CNVnator_v0.3/src/ ]
