Gap5 is very useful if you want to have a quick look at reads aligned to a part of a genome assembly. For example, say we have our read-to-genome stored in a BAM file 'my.bam', then we can extract reads aligning to positions 11000-13000 of scaffold 'myscaffold' by typing:
% samtools index my.bam my.bam.bai
% samtools view my.bam myscaffold:11000-13000 > my.sam
The first line indexes the BAM file so we can pull subsets of data from it. The second line makes a sam file my.sam containing the reads aligning to positions 11000-13000 of 'myscaffold'.
We can then make a Gap5 database file out of the SAM file my.sam by typing:
% /Users/alc/Documents/Software/Staden2.0.0b9/bin/tg_index my.sam
where /Users/alc/Documents/Software/Staden2.0.0b9/bin/tg_index is the path to the 'tg_index' program within the installed Staden package (note that this path may differ on your computer, depending on where you installed the Staden package).
This makes files my.0.g5d and my.0.g5x, which are the Gap5 database files.
To start up Gap5, you double-click on the Gap5 icon within the installed Staden package (eg. in directory 'Staden2.0.0b9'). This starts up Gap5, and brings up the main window.
You can select 'my.0.g5d' by going to the File menu, and choosing 'Open' and then choosing the my.0.g5.d file. This will bring up the 'Contig selector' which has your contigs (in this case just one contig 'myscaffold'):
In this case, there are quite few reads, so they are hard to distinguish from the white horizontal line.
To view the alignments of individual reads, click on the white horizontal line. This brings up a view of the alignments of individual reads:
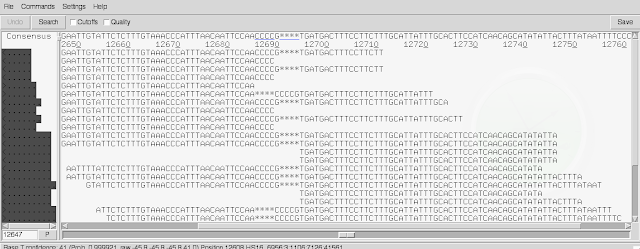
There are two scroll-bars across the bottom. The top one lets you move shorter distances, and the bottom one lets you move large distances. In the line at the top of the screen you can see the consensus sequence inferred for the genome sequence by Gap5, based on the read alignments.
By default, soft-clipped bases are not shown. If you tick the 'Cutoffs' box at the top of the window, they will be shown in pale grey.
Another useful thing to know is that by default, Gap5 gives 'padded coordinates', meaning that extra bases are inferred in the genome sequence (based on the reads) that were not in the original fasta file of scaffolds that the reads were aligned to, to make BAM file. In other words, in some cases, all the reads aligning at a position have an extra base that wasn't in the original fasta file, so this gives an extra base position in the consensus inferred by Gap5 at the top of the window. If you want to see the original reference genome coordinates (ie. coordinates with respect to the original fasta file of scaffolds) instead of the padded coordinates, go to the 'Settings' menu, and choose 'Reference coordinates'.
Thanks to Alan Tracey for introducing me to Gap5, a very nice tool.



No comments:
Post a Comment