For my data, I have a set of species in which I have drug targets; each drug target is present/absent in each species of my species set. I also have a species tree for the species, and have a similarity matrix with a similarity value for each pair of drug molecules.
I want to make a heatmap with the drugs on one axis, and species on the other axis, showing the presence/absence of drugs in the heatmap cells as dark/light squares. I also want to show the species tree beside the species, and a dendrogram based on drug similarities beside the drugs axis.
So now for some fun fiddling with R...
Here's how I did it (with first some notes on clustering, to remind myself):
Hierarchical clustering
Hierarchical clustering iteratively:
(i) identifies the closest pair of items/clusters
(ii) merges them into a new cluster
(iii) computes the distance between the new cluster and all the other items/clusters
It stops when all the items/clusters have been merged.
You need to define a method of calculating distances between two items, or between an item and a cluster. In single-linkage clustering, the distance between two clusters is the distance between the closest pair of items, for pairs where the two items belong to two different clusters. In average-linkage clustering the distance between two clusters is the average distance between pairs of items, for pairs where the two items belong to two different clusters.
By recording the order in which items/clusters were merged, the algorithm produces a tree. The lengths of vertical branches are proportional to the distances between clusters. After choosing how many clusters we want, we can 'cut' the tree at a certain height. This creates a hiearchy of clusters from small to big, depending on where you cut.
K-means clustering
An alternative to hierarchical clustering is K-means clustering. K-means clustering attempts to find the centres of clusters in the data. It aims to partition the items into k clusters, to minimise the sum of within-cluster distances. The user must specify how many clusters to find.
Clusters are defined by k points: 'centres'. An item belongs to the group defined by the nearest centre.
It starts off with a random choice of k centres. Then k initial clusters are created by assigning each gene to the nearest centre. It then replaces the k centres by the means (centroids) of the clusters.
Then we iterate again: each gene is then put into the cluster defined by the nearest centre. The algorithm then replaces the centres by the centroids of these new clusters.
Then we iterate again...and so on.
The algorithm iterates until it converges, ie. new clusters hardly differ from the previous clusters. An advantage is that we can plot the centroids to represent the clusters in a plot.
Reading in my similarity data for drugs
My data was a text file with this format: (tab-delimited)
CHEMBL923 CHEMBL1201269 CHEMBL415
CHEMBL923 1.00 0.63 0.22
CHEMBL1201269 0.63 1.00 0.89
CHEMBL415 0.22 0.89 1.00
To read it into R:
> mydata <- read.table("Compara_pairwise_sim.txt",header=TRUE,row.names=1)
(Note: skip=1 skips the first line of the file, row.names=1 uses the first column as the row names. colnames(mydata) <- rownames(mydata) would set the column names to be the same as row names.)
The values in my input file are similarities, scaled from 0-1, so I wanted to convert them to distances scaled from 0-1:
> mydata2 <- 1 - mydata
> mydist <- as.dist(mydata2)
This gives me a distance matrix 'mydist' for the drugs, which can be used as input for hierarchical clustering (below).
Building a dendrogram of drug clusters (to use later beside my heatmap), using hierarchical clustering
In R you can do K-means clustering using the 'kmeans' function, but here I'm going to use hierarchical clustering for my drugs. However, if you wanted to use K-means clustering you would type something like this, to find 5 clusters:
> drugclusters2 <- kmeans(mydist, 5)
See http://www.statmethods.net/advstats/cluster.html for some more advice.
For hierarchical clustering, you can do single-linkage or average-linkage clustering in R using the "hclust" function, with the method="single" or method="average" option. However, on http://www.statmethods.net/advstats/cluster.html, it suggests it is better to perform hierarchical clustering using the Ward's method (method="ward.D"), it says Ward's minimum variance method aims at finding compact, spherical clusters:
> drugclusters <- hclust(mydist, method="ward.D")
This gives me a clustering of my drugs.
Convert this to an R 'dendrogram' object: (we will use this to make the heatmap below)
> drugclusters_dendrogram <- as.dendrogram(drugclusters)
Display a dendrogram of the drug clusters, with the drug names in small text:
> plot(drugclusters, cex=0.5)
Cut the tree into 5 clusters:
> groups <- cutree(drugclusters, k=5)
Note: I once got an error message about 'the 'height' component of 'tree' is not sorted (increasingly)', but found a suggested solution to it here.
Draw the dendrogram with red borders around the 5 clusters:
> rect.hclust(drugclusters, k=5, border="red")
Read in my species tree, and make it into a dendrogram (to use later beside my heatmap)
For a heatmap, we need two dendrograms, one to use on the x-axis (eg. our dendrogram of drugs drugclusters above), and one to go on the y-axis (which I want to be my species tree).
I already have a species tree, so first need to convert the species tree to a dendrogram object in R:
(starting with a Newick-format species tree that has branch-lengths, but no bootstrap values)
> library("ape")
> species_tree <- read.tree("species_tree.nhx")
Use 'cronos' to convert the phylogenetic tree (with branch-lengths as substitutions/site) to a chronogram:
> chronogram <- chronos(species_tree)
Convert the chronogram to a dendrogram to use in a heatmap:
> species_tree_dendorgram <- as.dendrogram(as.hclust.phylo(dendrogram))
(Thanks to my former colleague Diogo Ribeiro for the details of how to do this.)
Read in the drug target presence/absence data to use in my heatmap
I have a tab-delimited file like this, with 1 for drug target presence and 0 for drug target absence:
species1 species2 species3...
drug1 1 0 1...
drug2 1 1 0...
I can read it into R like this:
> mydata = read.table("heatmap_data",header=TRUE)
Heatmap : step 1 : load libraries and set up data for ggplot2
I used ggplot2, and followed the protocol on http://stackoverflow.com :
> library("ggplot2")
> library(ggdendro,lib="~alc/R/library")
[Note to self: I had to install ggdendro: see here for how]
> library("reshape2")
Convert the data on drug target presence/absence to a matrix:
> mydata.matrix <- as.matrix(mydata)
For the columns of 'mydata' (the species), we will use the species dendrogram, ie. 'species_tree_dendrogram' (see above).
> species.ord <- order.dendrogram(species_tree_dendrogram)
For the rows of 'mydata' (the drug), we will use the drugs dendrogram, ie. "drugclusters_dendrogram" (see above).
> drugs.ord <- order.dendrogram(drugclusters_dendrogram)
Reorder the original data using the ordering of the columns and rows in the dendrograms:
> xx <- as.matrix(mydata[drugs.ord,species.ord])
Get the names of the drugs and species:
> xx_names <- attr(xx, "dimnames")
Convert it to a data frame:
> df <- as.data.frame(xx)
Set the column names of the data frame to be the species names:
> colnames(df) <- xx_names[[2]]
Make another column with the drug name:
> df$drug <- xx_names[[1]]
Rearrange the data:
> df$drug <- with(df, factor(drug, levels=drug, ordered=TRUE))
Now put the data in a nice format using 'melt' (it stacks a set of columns into a single column of data):
> mdf <- melt(df, id.vars="drug")
Heatmap : step 2 : extract dendrogram data and create the plots
> ddata_drugs <- dendro_data(drugclusters_dendrogram)
> ddata_species <- dendro_data(species_tree_dendrogram)
Set up a blank theme:
> theme_none <- theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.title.x = element_text(colour=NA),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.line = element_blank(),axis.ticks = element_blank())
> base_size <- 9
> p1 <- ggplot(mdf, aes(x=variable, y=drug)) + geom_tile(aes(fill=value)) + scale_fill_gradient2(low="white", high="steelblue") + theme_grey(base_size = base_size) + labs(x = "", y = "") + theme(axis.text.x = element_text(size = base_size * 1.2, angle = 330, hjust = 0, colour = "black")) + theme(axis.text.y = element_text(size = base_size * 0.2, colour = "black"))
Notes:
theme_grey(base_size=9) : sets the axis label text to size 'basesize', and colour grey.
labs(x = "", y="") : removes x-axis and y-axis labels from the plot.
Create plot components: (ii) dendrogram of species:
> p2 <- ggplot(segment(ddata_species)) + geom_segment(aes(x=x, y=y, xend=xend, yend=yend)) + theme_none + theme(axis.title.x=element_blank())
Create plot components: (iii) dendrogram of drugs:
> p3 <- ggplot(segment(ddata_drugs)) + geom_segment(aes(x=x, y=y, xend=xend, yend=yend)) + coord_flip() + theme_none
We now use grid graphics and some manual alignment to position the three plots on the page:
> library("grid")
> grid.newpage()
[Note that I needed to fiddle to vary the x-scale, y-scale, x-start-position, y-start-position in viewport to get the right size for the dendrogram below:]
> print(p2, vp=viewport(0.775, 0.2, x=0.385, y=0.9))
> print(p3, vp=viewport(0.2, 0.725, x=0.88, y=0.47))
> print(p1, vp=viewport(0.8, 0.8, x=0.4, y=0.4))
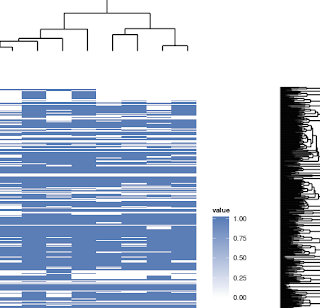
Part of my heatmap:
Nice webpages on using R for making heatmaps:
'Learning R' blogpost
Stackoverflow page on making a heatmap with ggplot2, showing dendrograms

No comments:
Post a Comment